How will Synthetic Biology and Conservation Shape the Future of Nature? Last month I was privileged to take part in a meeting organized by The Wildlife Conservation Society to consider that question. Here is the framing paper (PDF), of which I am a co-author. There will be a follow-up paper in the coming months. I am still mulling over what I think happened during the meeting, and below are a few observations that I have managed to settle on so far. Others have written their own accounts. Here is a summary from Julie Gould, riffing on an offer that Paul Freemont made to conservation biologists at the close of the meeting, "The Open Door". Ed Gillespie has a lovely, must-read take on Pandora's Box, cane toads, and Emily Dickenson, "Hope is the thing with feathers". Cristian Samper, the new head of the Wildlife Conservation Society was ultimately quite enthusiastic: Jim Thomas of ETC, unsurprisingly, not so much.
The meeting venue was movie set-like Cambridge. My journey took me through King's Cross, with its requisite mock-up of a luggage trolley passing through the wall at platform nine and three-quarters. So I am tempted to style parts of the meeting as a confrontation between a boyish protagonist trying to save the world and He Who Must Not Be Named. But my experience at the meeting was that not everyone was able to laugh at a little tension-relieving humor, or even to recognize that humor. Thus the title of this post is as much as I will give in temptation.
How Can SB and CB Collaborate?
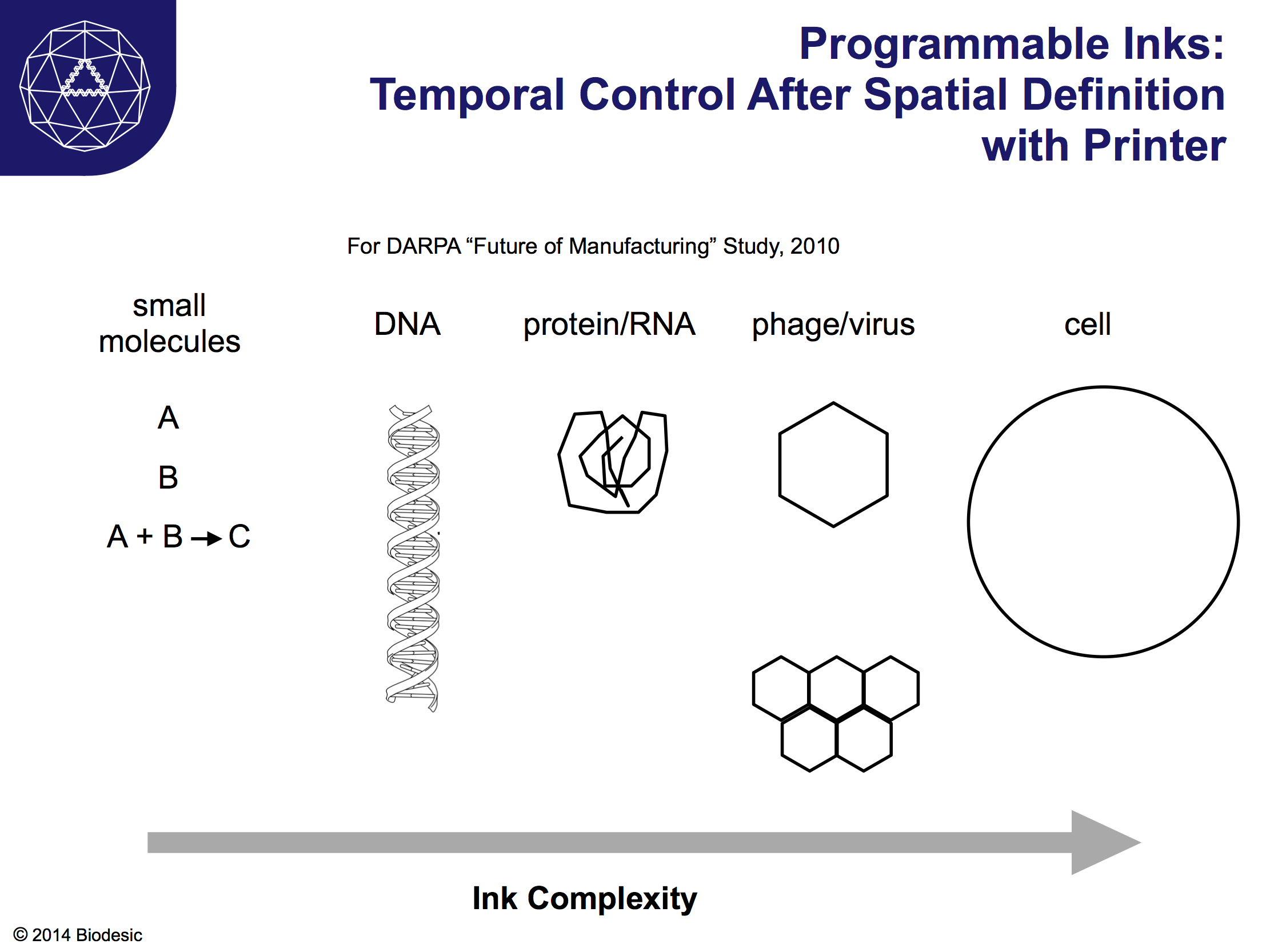
I'll start with an opportunity that emerged during the week, exactly the sort of thing you hope would come from introducing two disciplines to each other. What if synthetic biology could be used as a tool to aid in conservation efforts, say to buttress biodiversity against threats? If the ongoing, astonishing loss of species were an insufficient motivation to think about this possibility, now some species that humans explicitly rely upon economically are under threat. Synthetic biology might - might! - be able to offer help in the form of engineering species to be more robust in the face of a changing environment, such as enabling corals to cope with increases in water temperature and acidity, or it perhaps via intervening in a host-prey relationship, such as that between bats and white-nose disease or between bees and their mites and viruses.
The first thing to say here is that if the plight of various species can be improved through changes in human behavior then we should by all means work toward that end. The simpler solution is usually the better solution. For example, it might be a good idea to stop using those pesticides and antibiotics that appear to create more problems than they solve when introduced into the environment. Moreover, at the level of the environment and the economy, technological fixes are probably best reserved until we try changes in human behavior. After all, we've mucked up such fixes quite a few times already. (All together now: "Cane Toad Blues".) But what if the damage is too far along and cannot be addressed by changes in behavior? We should at least consider the possibility that a technological fix might be worth a go, if for no other reason that to figure out how to create a back up plan. Given the time scales involved in manipulating complex organisms, exploring the option of a back-up plan means getting started early. It also means thoughtfully considering which interventions would be most appropriate and urgent, where part of the evaluation should probably involve asking whether changes in human behavior are likely to have any effect. In some cases, a technical solution is likely to be our only chance.
First up: corals. We heard from Stanford's Steve Palumbi on work to understand the effects of climate change on corals in the South Pacific. Temperature and acidity - two parameters already set on long term changes - are already affecting coral health around the globe. But it turns out that in the lab some corals can handle remarkably difficult environmental conditions. What if we could isolate the relevant genetic circuits and, if necessary, transplant them into other species, or turn them on if they are already widespread? My understanding of Professor Palumbi's talk is that it is not yet clear why some corals have the pathway turned on and some do not. So, first up, a bunch of genetics, molecular biology, and field biology to figure out why the corals do what they do. After that, if necessary, it seems that it would be worth exploring whether other coral species can be modified to use the relevant pathways. Corals are immensely important for the health of both natural ecosystems and human economies; we should have a back-up plan, and synthetic biology could certainly contribute.
Next up: bats. Bats are unsung partners of human agriculture, and they contribute an estimated $23 billion annually to U.S. farmers by eating insects and pollinating various plants. Here is nice summary article from The Atlantic by Stephanie Gruner Buckely on the impact upon North American bats of white nose syndrome. The syndrome, caused by a fungus evidently imported from Europe, has already killed so many bats that we may see an impact on agriculture as soon as this year. European bats are resistant to the fungus, so one option would be to try to introduce the appropriate genes into North American bats via standard breeding. However, bats breed very slowly, usually only having one pup a year, and only 5 or so pups in a lifetime. Given the mortality rate due to white nose syndrome, this suggests breeding is probably too slow to be useful in conservation efforts. What if synthetic biology could be used to intervene in some way, either to directly attack the non-native fungus or to interfere with its attack on bats. Obviously this would be a hard problem to take on, but both biodiversity and human welfare would be improved by making progress here.
And now: bees. If you eat, you rely on honeybees. Due to a variety of causes, bee populations have fallen to the point where food crops are in jeopardy. Entomologist Dennis vanEngelstorp, quoted in Wired, warns "We're getting closer and closer to the point where we don't have enough bees in this country to meet pollination demands. If we want to grow fruits and nuts and berries, this is important. One in every three bites [of food consumed in the U.S.] is directly or indirectly pollinated by bees." Have a look at the Wired article for a summary of the constellation of causes of Colony Collapse Disorder, or CCD -- they are multifold and interlocking. Obviously, the first thing to do is to stop making the problem worse; Europe has banned a class of pesticide that is exceptionally hard on honeybees, though the various sides in this debate continue to argue about whether that will make any difference. This change in human behavior may have some impact, but most experts agree we need to do more. Efforts are underway to breed bees that are resistant to both pesticides and to particular mites that prey on bees and that transmit viruses between bees. Applying synthetic biology here might be the hardest task of all, given the complexity of the problem. Should synthetic biologists focus on boosting apian immune systems? Should they focus on the mite? Apian viruses? It sounds very difficult. But with such a large fraction of our food supply dependent upon healthy bees, it also seems pretty clear that we should be working on all fronts to sort out potential solutions.
A Bit of Good News
Finally, a problem synthetic biologists are already working to solve: malaria. The meeting was fortunate to hear directly from Jay Keasling. Keasling presented progress on a variety of fronts, but the most striking was his announcement that Sanofi-Aventis has produced substantially more artemisinin this year than planned, marking real progress in producing the best malaria drug extant using synthetic biology rather than by purifying it from plants. Moreover, he announced that Sanofi and OneWorldHealth are likely to take over the entire world production of artemisinin. The original funding deal between The Gates Foundation, OneWorldHealth, Amyris, and Sanofi required selling at cost. The collaboration has worked very hard at bringing the price down, and now it appears that they can simply outcompete the for-profit pricing monopoly.
The stated goal of this effort is to reduce the cost of malaria drugs and provide inexpensive cures to the many millions of people who suffer from malaria annually. Currently, the global supply fluctuates, as, consequently, do prices, which are often well above what those afflicted can pay. A stable, high volume source of the drug would reduce prices and also reduce the ability of middle-men to sell doctored, diluted, or mis-formulated artemisinin, all of which are contributing to a rise of resistant pathogens.
There is a potential downside to this project. If Sanofi and OneWorldHealth do corner the market on artemisinin, then farmers who currently grow artemisia will no longer have that option, at least for supplying the artemisinin market. That might be a bad thing, so we should at least ask the question of whether the world is a better place with artemisinin production done in vats or derived from plants. This question can be broken into two pieces: 1) what is best for the farmers? and 2) what is best for malaria sufferers? It turns out these questions have the same answer.
There is no question that people who suffer from malaria will be better off with artemisinin produced in yeast by Sanofi. Malaria is a debilitating disease that causes pain, potentially death, and economic hardship. The best estimates are that countries in which malaria is endemic suffer a hit to GDP growth of 1.3% annually compared to non-malarious countries. Over just a few years this yearly penalty swamps all the foreign aid those countries receive; I've previously argued that eliminating malaria would be the biggest humanitarian achievement in history and would make the world a much safer place. Farmers in malarious countries are the worst hit, because the disease prevents them from getting into the fields to work. I clashed in public over this with Jim Thomas around our respective testimonies in front of the Presidential Bioethics Commission a couple of years ago. Quoting myself briefly from the relevant blog post,
The human cost of not producing inexpensive artemisinin in vats is astronomical. If reducing the burden of malaria around the world on almost 2 billion people might harm "a few thousand" farmers, then we should make sure those farmers can make a living growing some other crop. We can solve both problems. ...Just one year of 1.3% GDP growth recovered by reducing (eliminating?) the impact of malaria would more than offset paying wormwood farmers to grow something else. There is really no argument to do anything else.
For a bit more background on artemisinin supply and pricing, and upon the apparent cartel in control of pricing both the drug and the crop, see this piece in Nature last month by Mark Peplow. I was surprised to learn that that the price of artemisia is set by a small group that controls production of the drug. This group, unsurprisingly, is unhappy that they may lose control of the market for artemisinin to a non-profit coalition whose goal is to eliminate the disease. Have a look at the chart titled "The Cost of Progress", which reveals substantial price fluctuations, to which I will return below.
Mr. Thomas responded to Keasling's announcement in Cambridge with a broadside in the Guardian UK against Keasling and synthetic biology more generally. Mr. Thomas is always quick to shout "What about the farmers?" Yet he is rather less apt to offer actual analysis of what farmers actually gain, or lose, by planting artemisia.
The core of the problem for farmers is in that chart from Nature, which shows that artemisinin has fluctuated in price by a factor of 3 over the last decade. Those fluctuations are bad for both farmers and malaria sufferers; farmers have a hard time knowing whether it makes economic sense to plant artemisia, which subsequently means shortages if farmers don't plant enough. Shortages mean price spikes, which causes more farmers to plant, which results in oversupply, which causes the price to plunge, etc. You'll notice that Mr. Thomas asserts that farmers know best, but he never himself descends to the level of looking at actual numbers, and whether farmers benefit by growing artemisia. The numbers are quite revealing.
Eyeballing "The Cost of Progress" chart, it looks like artemisia has been below the $400/kg level for about half the last 10 years. To be honest, there isn't enough data on the chart to make firm conclusions, but it does look like the most stable price level is around $350/kg, with rapid and large price spikes up to about $1000/kg. Farmers who time their planting right will probably do well; those who are less lucky will make much less on the crop. So it goes with all farming, unfortunately, as I am sure Mr. Thomas would agree.
During his talk, Keasling put up a chart I hadn't seen before, which showed predicted farmer revenues for a variety of crops. The chart is below; it makes clear that farmers will have substantially higher revenues planting crops other than artemisia at prices at or below $400/kg.